The Hesketh Lab @ Dalhousie University
Lysosomal control of cellular anabolic-catabolic balance

My name is Geoffrey Hesketh. I am a molecular cell biologist, fascinated by how lysosomes control the balance between anabolic and catabolic pathways in cells. Specifically, I study how nutrients are sensed on the surfaces of lysosomal system organelles and how this is coupled to the activation state of mTORC1, a master growth regulator. mTORC1 in turn broadly controls cellular anabolism (i.e., macromolecular biosynthesis) and catabolism (i.e., macromolecular breakdown). These pathways are central to the pathogenesis of human diseases including mutant Ras-driven cancers (e.g., pancreatic cancer), neurodegeneration, and infectious diseases. In pursuit of these questions I employ proximity-dependent biotinylation and mass spectrometry (e.g., BioID), CRISPR-Cas9 gene editing, and a variety of classical molecular cell biology techniques.
I am currently an Assistant Professor in the Department of Biochemistry & Molecular Biology in the Faculty of Medicine at Dalhousie University in Halifax, Nova Scotia, Canada. I was a postdoctoral fellow in the laboratory of Dr. Anne-Claude Gingras at the Lunenfeld-Tanenbaum Research Institute (LTRI) in Toronto, Ontario. I previously completed a postdoctoral fellowship in the laboratories of Prof. Paul Luzio and Prof. David Owen at the Cambridge Institute for Medical Research (CIMR) at the University of Cambridge in Cambridge, UK.
I completed my PhD under the supervision of Dr. Jennifer Van Eyk and Dr. Gordon Tomaselli in the Department of Biological Chemistry at the Johns Hopkins University School of Medicine in Baltimore, Maryland.
Link to my CV here.
Link to a detailed research proposal here.
Link to my recent publication in the journal Science
Research
The lysosomal system and mTORC1 activation by nutrients
![The lysosomal system and mTORC1 activation by nutrients]()
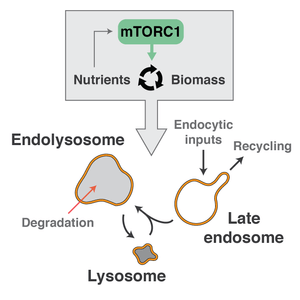
The lysosomal system consists of late endosomes, lysosomes, and the product of their fusion, endolysosomes. Late endosomes receive traffic downstream of endocytic pathways (e.g., macropinocytosis and receptor-mediated endocytosis) and engage in a variety of recycling pathways (e.g., retromer pathway). Storage lysosomes are effectively "sacs" of hydrolytic enzymes, which become active upon fusion with late endosomes. This results in degradation of the endolysosomal luminal contents and the liberation of nutrients from the breakdown of macromolecules. Soluble nutrients (e.g., amino acids) can be transported directly to the cytoplasm, whereas insoluble nutrients (e.g., lipids) are transferred from endolysosomes to other organelles via membrane contact sites and lipid transfer proteins.
The mechanistic Target of Rapamycin (mTOR) kinase complex 1 (mTORC1) is a master regulator of nutrient-dependent cellular processes. Nutrients, acting through a variety of distinct nutrient sensing mechanisms, control the activation state of mTORC1. In response, mTORC1 broadly controls the balance between cellular anabolism (i.e., creation of biomass from nutrients) and cellular catabolism (i.e., the breakdown of biomass to generate nutrients) according to nutrient availability. Importantly, nutrient sensing and activation of mTORC1 occurs on the surfaces of lysosomal system organelles.
Please see my recent publication in the journal Science identifying a novel mechanism of mTORC1 activation by endolysosome-derived amino acids.
Spatio-temporal proteomics of the lysosomal system using BioID
![Spatio-temporal proteomics of the lysosomal system using BioID]()
Proximity-dependent biotinylation with biotin protein ligases and mass spectrometry (referred to here as BioID) is a powerful technique allowing the identifcation of proximity-dependent associations within protein communities in live cells. Please see this recent review from the lab for a detailed explanation of proximity-dependent biotinylation techniques. Using BioID on a large scale, combined with advanced data analysis techniques, the lab created a BioID proximity map of the HEK293 proteome called the Human Cell Map. This project was recently published in the journal Nature (C. Go and J. Knight co-first authors).
I have used BioID to profile greater than 100 proteins involved in lysosomal system membrane trafficking and mTORC1 signalling. This work has resulted in a rich dataset from which multiple hypotheses will be tested functionally. Importantly, certain baits behave as "organelle sensors" and, when combined with a technique I developed called "Knockout BioID" (KO-BioID), provide detailed mechanistic insight into trafficking and signalling events occurring on organelle surfaces. KO-BioID using the lysosomal system SNARE proteins VAMP7 and VAMP8 was the catalyst that led to my recent publication in Science.
In my independent career I will continue to use BioID as a central discovery tool, gaining mechanistic insight into lysosomal control of cellular anabolic and catabolic pathways.
Projects will fall under two broad aims, further outlined in the following sections.
i) Investigate mechanisms by which lysosome-driven growth pathways fuel highly lethal mutant Ras-driven cancers.
ii) Investigate the role of the lysosomal system in orchestrating cellular nutrient biology.i) Lysosome-driven growth in Ras-driven cancers
![i) Lysosome-driven growth in Ras-driven cancers]()
Oncogenic Ras-driven cancers, such as pancreatic cancer, are highly lethal with virtually no targeted therapies available. A major consequence of oncogenic Ras expression in cells is the constitutive activation of macropinocytosis (cellular "drinking"), allowing massive uptake and endolysosomal degradation of macromolecule-rich extracellular fluid. These lysosome-derived nutrients activate mTORC1 through a novel mechanism, and can fuel cancer cell growth in the nutrient-deprived tumour microenvironment. I will use a combination of BioID, genome-wide CRISPR screens, and hypothesis-driven cell biology to investigate:
a) Mechanisms of mTORC1 activation by lysosome-derived nutrients and how this impacts function of the lysosomal system.
b) The contribution of lysosome-derived nutrients to Ras-driven cancer pathobiology.ii) Lysosomal control of cellular nutrient biology
![ii) Lysosomal control of cellular nutrient biology]()
My extensive BioID screen investigating lysosomal system membrane trafficking and signalling has allowed the generation of multiple hypotheses related to lysosomal control of cellular nutrient biology. Initial projects will investigate:
a) Mechanistic roles for the Rab7-coordinated HOPS and Retromer complexes in controlling nutrient-dependent late endosome-lysosome fusion and recycling.
b) Mechanistic basis for membrane contacts and lipid transfer proteins in controling nutrient (e.g., cholesterol) acquisition, cellular distribution, and signalling.
c) Mechanisms of nutrient-dependent regulation of gene expression.
Collectively, through these studies I aim to illuminate how defects in lysosomal control of cellular anabolic-catabolic balance cause human disease.
Opportunities
Potential students, please get in touch by sending me an email.
Contact
Geoffrey G. Hesketh PhD
Geoffrey.Hesketh@Dal.ca
Department of Biochemistry & Molecular Biology
Dalhousie University
Halifax, NS, Canada
tel: 902-494-3821